Neurosynth
Your meta-analytical field guide in the brain's functional wilderness
A meta-analysis is a statistical procedure that compiles and analyzes data from multiple independent studies with overlapping research themes to draw robust conclusions about a specific question. It is, essentially, a study of studies that typically takes months of hard work sifting through numerous publications that meet explicit inclusion/exclusion criteria to curate a large enough dataset that can be statistically dissected to reveal clear overall trends. Thanks to the magic of modern analytical tools, this tortuous process can be significantly streamlined—and in this issue of ZAGROSCIENCE, I will show you how to synthesize your own personalized brain meta-analysis in less than 30 seconds by using an online interface known as Neurosynth.
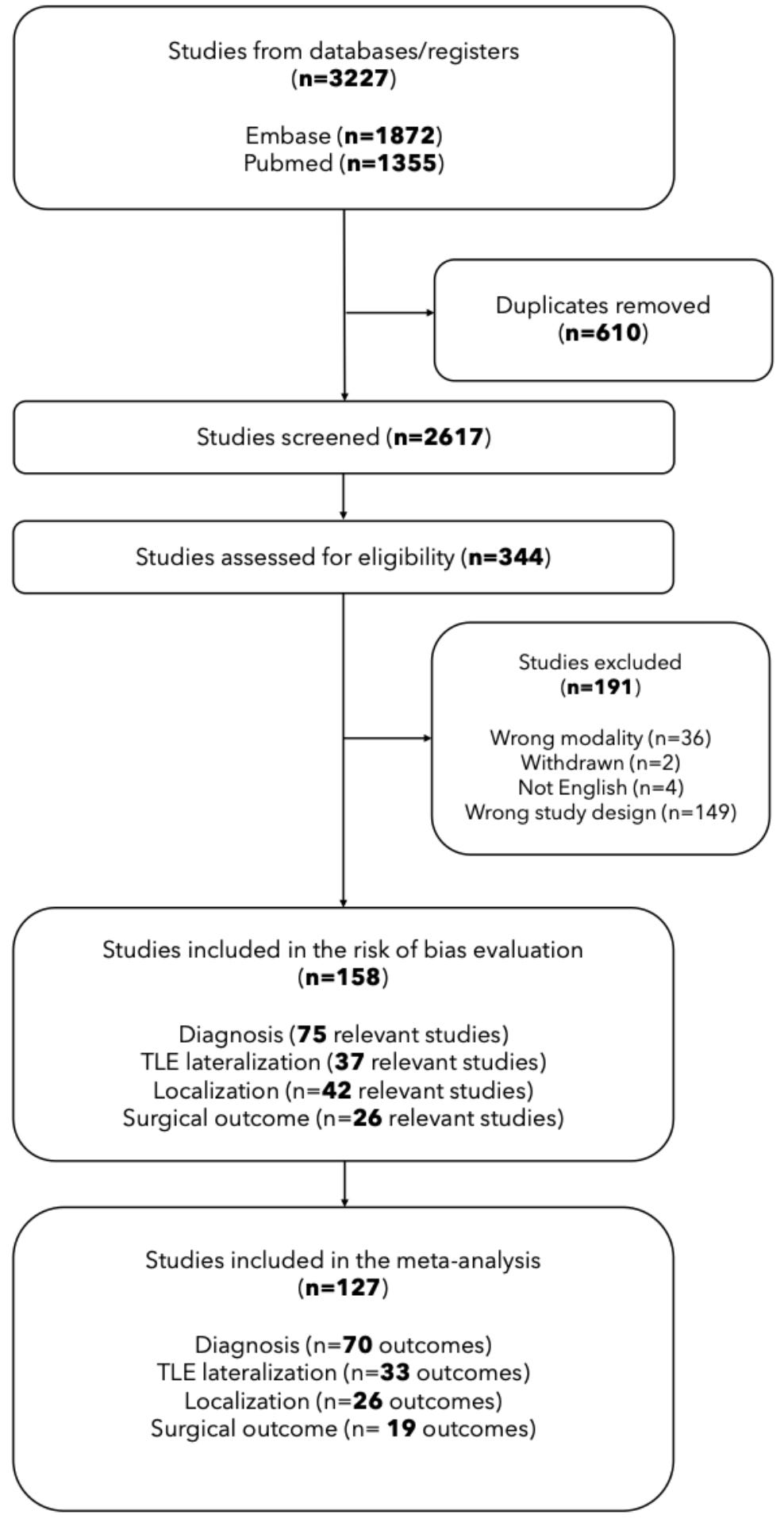
Before we get started, let us take a close look at a specific example of a meta-analysis to develop an intuition for how the whole process actually works. It just so happens that, last week, my colleagues at MICA (https://mica-mni.github.io/) released a preprint (Chen et al., 2025 PREPRINT) that examines the effectiveness of artificial intelligence (AI)/machine learning (ML) in aiding with the diagnosis and management of epilepsy. For those unfamiliar, a preprint is an openly available scientific publication that has not been peer-reviewed. It is a useful way for researchers to share their latest work with the scientific community for feedback while preparing the final version of the manuscript for academic vetting by independent experts, which will determine whether or not it will be officially published in a scholarly journal. Part systematic review and part meta-analysis, the authors of Chen et al., investigated the accuracy of AI/ML models applied to multimodal MRI data in predicting: (1) diagnostic status (i.e., epilepsy vs. no epilepsy), (2) disease lateralization (i.e., left vs. right hemisphere), (3) lesion localization (i.e., frontal lobe, hippocampus, parahippocampus, etc.), and (4) post-surgery prognosis (i.e., seizure-freedom vs. recurrence). Relevant studies were identified from PubMed, Medline, and Embase databases, ensuring that each one included a minimum of 10 patients and at least one of the following classifier evaluation metrics: sensitivity/specificity, positive predictive value/negative predictive value, F1 score, ROC AUC, or raw accuracy rate. Of the originally identified 3,227 studies, statistical analyses were conducted on the 127 that met inclusion/exclusion criteria, determining overall accuracy scores and assessing the impact of factors like cohort size and algorithm type on performance. Results showed that AI/ML models were 88% accurate in diagnosis, 90% accurate in lateralization, 82% accurate in localization, and 83% accurate in post-surgical outcome prediction. Even so, the authors pointed to significant bias in how studies recruited participants, determined predictors/outcome measures, and conducted analyses. Overall, the meta-analysis suggests that while AI holds immense promise for the identification and treatment of epilepsy, unlocking this potential will require rigorous, transparent, and trustworthy science.
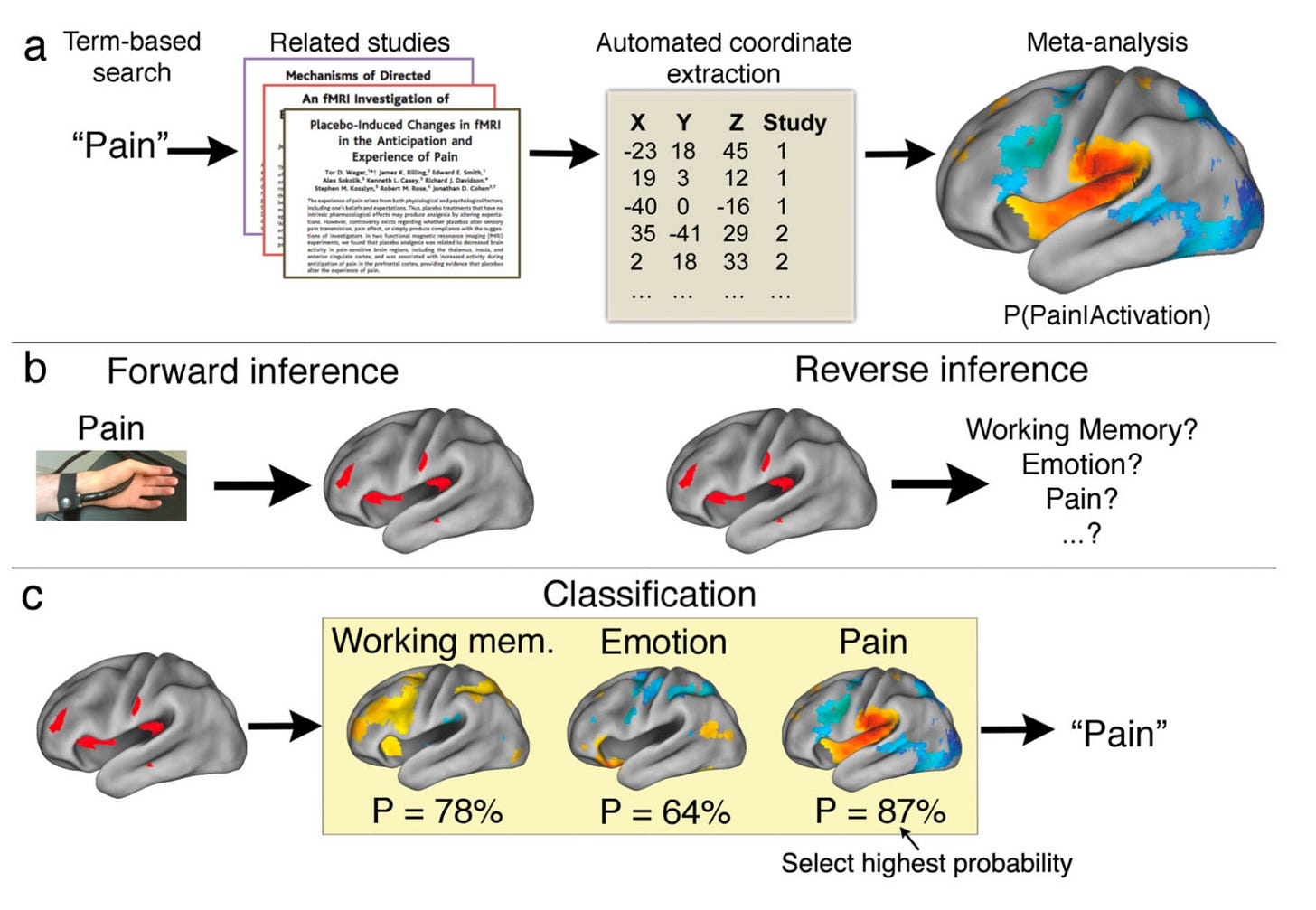
Let us now see how you can produce your own meta-analytical brain synthesis using an openly accessible platform that is widely used among network neuroscientists. In 2011, Yarkoni et al., (2011) introduced the world to NeuroSynth, an online toolbox that seamlessly maps between cognitive states and functional brain patterns by integrating machine learning, text mining, and meta-analysis. It leverages large-scale fMRI and textual data obtained from an extensive scientific corpus to automatically generate probabilistic brain maps determined by user-defined term-based queries (i.e., forward inference) or to classify previously derived cortical patterns into associated terms based on posterior probability maps for various other terms (i.e., reverse inference or decoding).
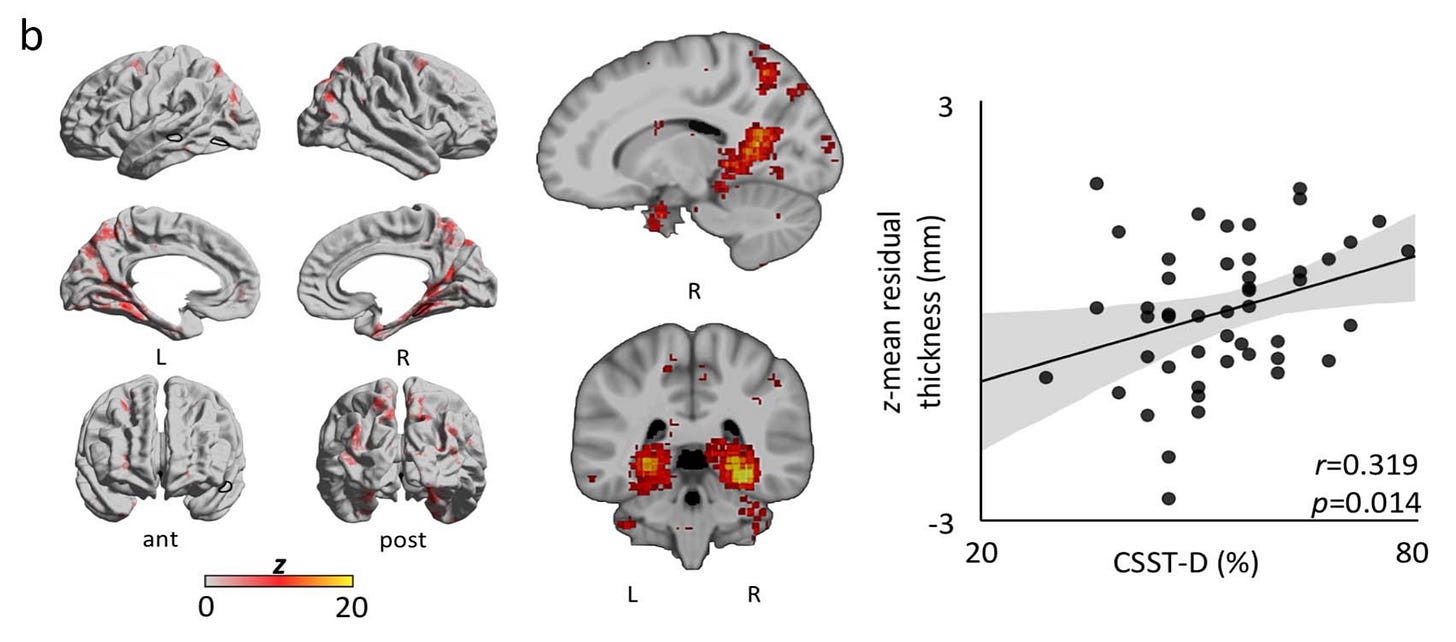
In Tavakol et al., (2021), my second doctoral publication, my co-authors and I implemented Neurosynth to analytically recapitulate 77 studies with a total 3,908 activations, resulting in a single cortical association map that we used to regionally constrain additional analyses.
The following is a step-by-step guide on how to conduct your own Neurosynth-based meta-analysis in less than 30 seconds:
Access Neurosynth by clicking this link: https://neurosynth.org/
Click the “Meta-analyses” option on the navigation menu at the top
Select “Terms”
Enter a query in the search field (i.e., “memory”)
Select your preferred search result from the options (i.e., “memory)
And voila! If you had chosen “memory” as both your query and search result, you would have automated a meta-analysis of 2,744 studies resulting in whole-brain activation patterns displayed on the MNI152 volumetric template, with clusters of finding in the medial temporal lobes (see screenshot below).
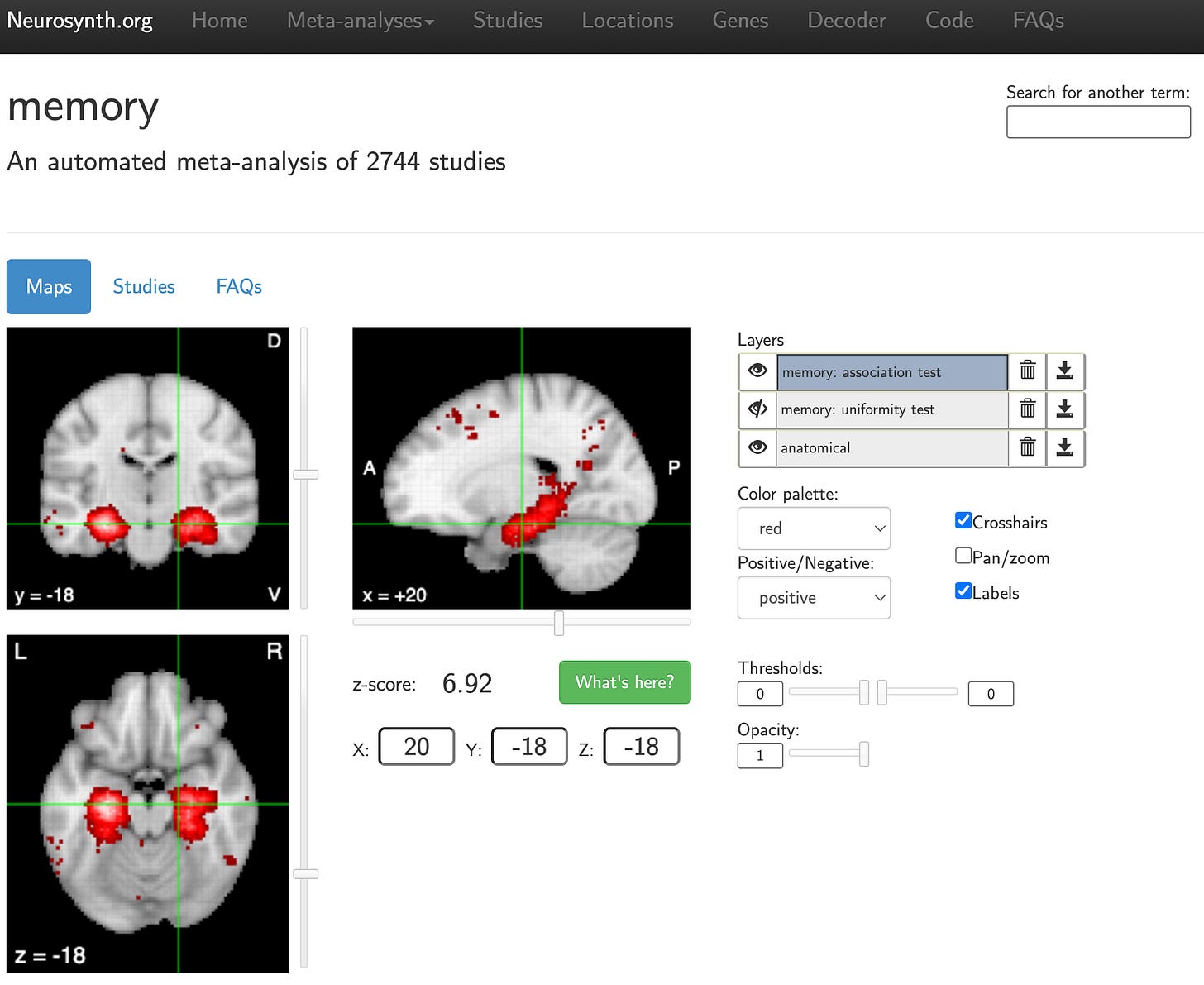
What makes Neurosynth such a useful tool is that, in addition to generating on the fly meta-analytical fMRI co-activation maps based on input terms, it can also operate in the reverse direction. Indeed, one can extract terms relating to cognitive processes by feeding into its algorithm previously derived statistical brain maps. In Vos de Wael et al., (2021), which I co-authored, we implemented Neurosynth to decode orthogonal patterns of structural connectivity in the human temporal lobe to provide some cognitive context to the underlying axes. We observed that principle gradients of structural organization in the temporal lobe recapitulate known variations in its roles pertaining to sensory function and cognitive processes (see figure below).
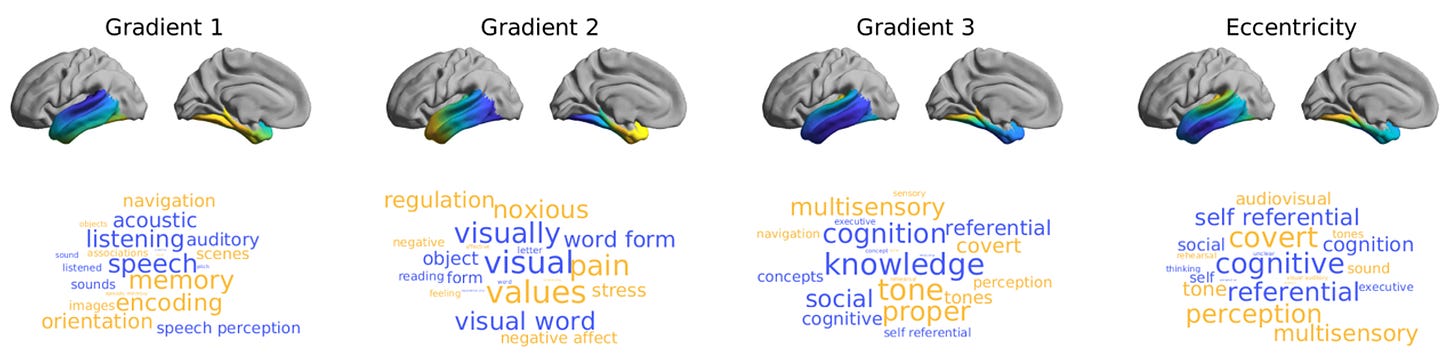
The wilderness of the brain is a landscape filled with unexpected twists and turns, and the job of the neuroscientist is to make sense of it all. A brain-based meta-analysis is a powerful statistical approach for scrutinizing a systematized dataset constructed from multiple sources to tease out informative overall trends. Tools like Neurosynth that streamline forward and reverse statistical inferencing of large-scale neural and textual data enable researchers to easily derive meta-analytical insights that contribute significantly to our understanding of what the brain is and how it works. And now that you know about the basics of Neurosynth, you can take part in this endeavour by conducting your own personalized neuroscientific syntheses. Enjoy!
References
Chen, J., Sahlas, E., Zhou, Y., Chen, N., Xie, J., Wadia, F., ... & Bernhardt, B. (2025). The use of Artificial Intelligence in Magnetic Resonance Imaging of Epilepsy: A Systematic Review and Meta-Analysis. bioRxiv, 2025-09.
Neurosynth. (n.d.). https://neurosynth.org/
Tavakol, S., Li, Q., Royer, J., Vos de Wael, R., Larivière, S., Lowe, A., ... & Bernhardt, B. (2021). A structure–function substrate of memory for spatial configurations in medial and lateral temporal cortices. Cerebral Cortex, 31(7), 3213-3225.
Vos de Wael, R., Royer, J., Tavakol, S., Wang, Y., Paquola, C., Benkarim, O., ... & Bernhardt, B. C. (2021). Structural connectivity gradients of the temporal lobe serve as multiscale axes of brain organization and cortical evolution. Cerebral cortex, 31(11), 5151-5164.
Yarkoni, T., Poldrack, R. A., Nichols, T. E., Van Essen, D. C., & Wager, T. D. (2011). Large-scale automated synthesis of human functional neuroimaging data. Nature methods, 8(8), 665-670.
Note: The author “Tavakol, S.” is Shahin Zagros (myself).